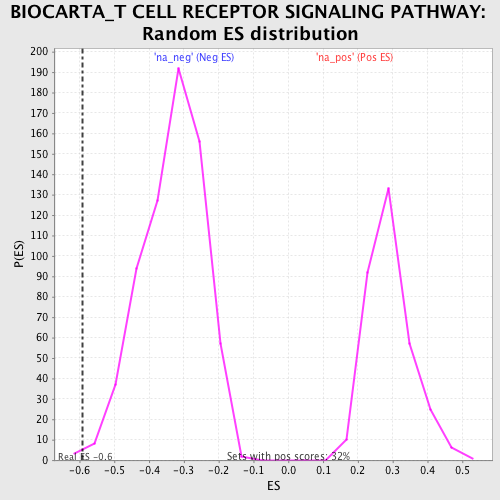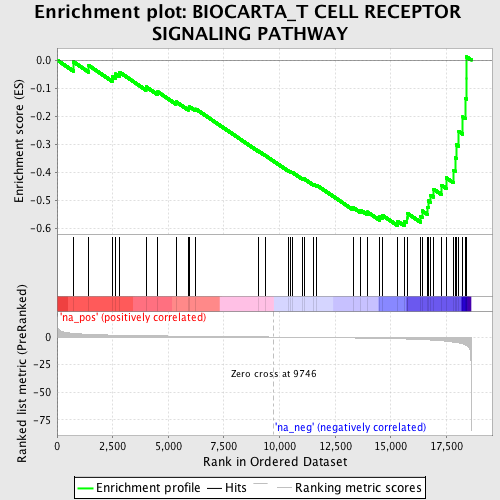
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | BIOCARTA_T CELL RECEPTOR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.5914742 |
| Normalized Enrichment Score (NES) | -1.7807237 |
| Nominal p-value | 0.00443787 |
| FDR q-value | 0.12873307 |
| FWER p-Value | 0.913 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GRB2 | 736 | 3.346 | -0.0065 | No | ||
| 2 | PTPRC | 1422 | 2.430 | -0.0193 | No | ||
| 3 | MAP2K4 | 2476 | 1.763 | -0.0585 | No | ||
| 4 | PLCG1 | 2622 | 1.699 | -0.0495 | No | ||
| 5 | PPP3CB | 2813 | 1.620 | -0.0437 | No | ||
| 6 | MAPK8 | 3997 | 1.205 | -0.0954 | No | ||
| 7 | ARHGAP6 | 4528 | 1.070 | -0.1134 | No | ||
| 8 | RAF1 | 5344 | 0.875 | -0.1486 | No | ||
| 9 | NCF2 | 5899 | 0.759 | -0.1709 | No | ||
| 10 | RAC1 | 5946 | 0.749 | -0.1660 | No | ||
| 11 | PIK3R1 | 6233 | 0.693 | -0.1745 | No | ||
| 12 | HRAS | 9039 | 0.147 | -0.3241 | No | ||
| 13 | MAP2K1 | 9344 | 0.087 | -0.3396 | No | ||
| 14 | PRKCA | 10410 | -0.150 | -0.3954 | No | ||
| 15 | JUN | 10501 | -0.169 | -0.3986 | No | ||
| 16 | SOS1 | 10560 | -0.179 | -0.4000 | No | ||
| 17 | ARHGAP1 | 11045 | -0.278 | -0.4233 | No | ||
| 18 | FOS | 11102 | -0.290 | -0.4234 | No | ||
| 19 | CAMK2B | 11527 | -0.379 | -0.4425 | No | ||
| 20 | ARHGAP5 | 11672 | -0.412 | -0.4462 | No | ||
| 21 | RALBP1 | 13313 | -0.815 | -0.5264 | No | ||
| 22 | PPP3CA | 13649 | -0.916 | -0.5354 | No | ||
| 23 | MAPK3 | 13948 | -1.009 | -0.5414 | No | ||
| 24 | PIK3CA | 14498 | -1.205 | -0.5590 | No | ||
| 25 | PPP3CC | 14611 | -1.241 | -0.5528 | No | ||
| 26 | ARFGAP1 | 15302 | -1.541 | -0.5747 | Yes | ||
| 27 | ZAP70 | 15615 | -1.707 | -0.5746 | Yes | ||
| 28 | ARFGAP3 | 15727 | -1.774 | -0.5630 | Yes | ||
| 29 | LAT | 15730 | -1.775 | -0.5455 | Yes | ||
| 30 | SHC1 | 16333 | -2.217 | -0.5559 | Yes | ||
| 31 | MAP3K1 | 16413 | -2.279 | -0.5376 | Yes | ||
| 32 | TRB@ | 16652 | -2.512 | -0.5256 | Yes | ||
| 33 | NFKBIA | 16685 | -2.540 | -0.5021 | Yes | ||
| 34 | PTPN7 | 16789 | -2.668 | -0.4812 | Yes | ||
| 35 | CD3D | 16927 | -2.841 | -0.4605 | Yes | ||
| 36 | LCK | 17279 | -3.355 | -0.4461 | Yes | ||
| 37 | RELA | 17490 | -3.802 | -0.4198 | Yes | ||
| 38 | CD3G | 17824 | -4.591 | -0.3922 | Yes | ||
| 39 | CD4 | 17903 | -4.846 | -0.3484 | Yes | ||
| 40 | VAV1 | 17943 | -4.944 | -0.3016 | Yes | ||
| 41 | FYN | 18056 | -5.334 | -0.2547 | Yes | ||
| 42 | CD247 | 18236 | -6.229 | -0.2027 | Yes | ||
| 43 | ARHGAP4 | 18360 | -7.246 | -0.1375 | Yes | ||
| 44 | NFATC1 | 18404 | -7.630 | -0.0643 | Yes | ||
| 45 | CD3E | 18405 | -7.633 | 0.0114 | Yes |